Overview
I am interested rigorous multiscale modeling for soft matter and biophysical systems with applications using large-scale computations. Ultimately, my goal is to use computational approaches to solve concrete soft matter and biomedical engineering problems through the use and development of data-driven mesoscopic models, high-performance softwares and massively parallel supercomputers. My research has been built around this central idea on three different fronts: 1) construction of mesoscopic models for amphiphilic and thermo-responsive polymers, 2) development of computational frameworks and high-performance simulators, and 3) application of large-scale mesoscopic simulations, as manifested in the projects below.
Self-Assembly of Function Materials
Understanding the mechanism of self-assembly is crucial for fabricating functional materials, e.g. controlled-release micro drug carriers, and promoting their adoption into real-world applications. To this end, I have investigated the dynamic self-assembly process of amphiphilic polymers in uneven temperature fields and within softly confined spaces with large-scale dissipative particle dynamics simulations.
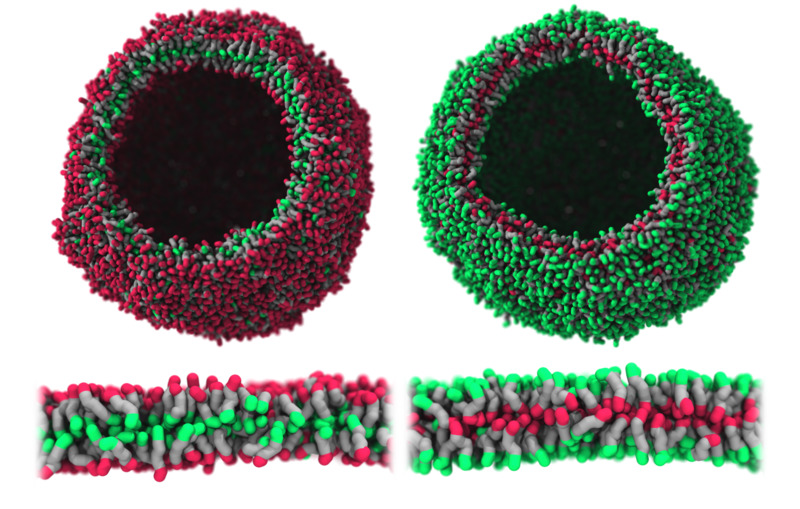
Using a new energy-conserving thermo-responsive polymer model, I discovered that bilayer membranes with dual thermoresponsivity exhibit a significant ratio of vertically translational molecular migration between the two leaflets. This type of behavior is distinct from the biological lipid bilayers. Our simulations of micelles traveling in micro-channels also reveal that thermoresponsive carriers exhibit anomalous hydrodynamic behavior at thermal interfaces in flow fields with uneven temperature distribution, an essential scenario to be considered for designing temperature-responsive drug delivery systems.

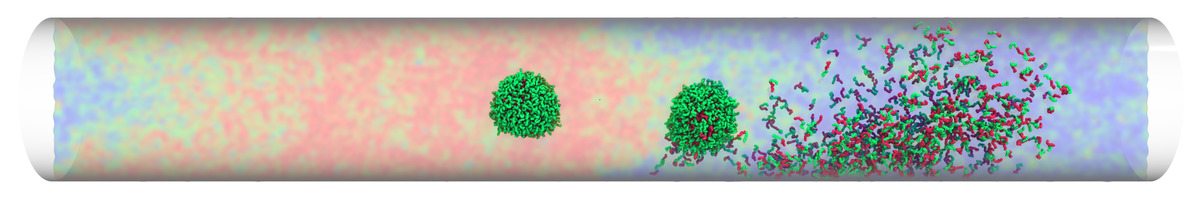
Micelles traveling in microtube: thermo-reponsove micelles may invert or break down when moving across thermal interfaces, while a non-responsive micelle remains inert.
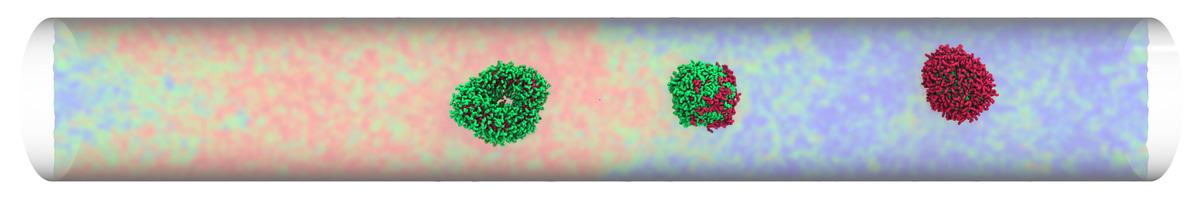
Multiblock copolymers with thermo-responsible end blocks can flip its inside and outside composition upon heating or cooling.
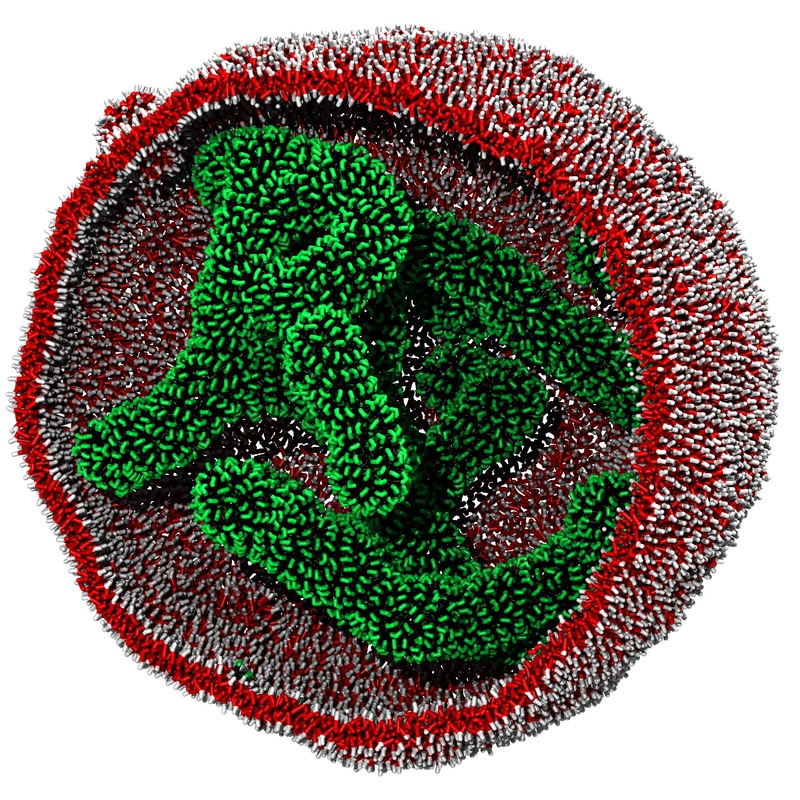
Self-assembly of amphiphilic molecules in another spherical vesicle which serves as a 'soft' confinement.
| [1] | Non-Equilibrium Dynamics of Vesicles and Micelles by Self-Assembly of Block Copolymers with Double Thermoresponsivity. Macromolecules, 49(7):2895-2903, 2016. |
| [2] | Mesoscale modeling of phase transition dynamics of thermoresponsive polymers. Chemical Communications, 51(55):11038-11040, 2015. |
| [3] | Large-scale dissipative particle dynamics simulations of self-assembled amphiphilic systems. Chemical Communications, 50(61):8306-8308, 2014. |
Multiscale Coupling
Concurrently coupled numerical simulations using heterogeneous solvers are powerful tools for modeling multiscale phenomena. However, major modifications to existing codes are often required to enable such simulations, posing significant difficulties in practice. I started the Multiscale Universal Interface (MUI) project with the aim to address the difficulty in implementing concurrently coupled multiscale simulations across a diverse array of solvers and methods by providing a very simple, yet powerful, set of interfaces for inter-solver communications. By introducing the concept of data points and data sampler, MUI gives users the freedom to choose their own spatial and temporal coupling scheme.
| [1] | Multiscale Universal Interface: A concurrent framework for coupling heterogeneous solvers. Journal of Computational Physics, 297:13-31, 2015. |
| [2] | Analysis of hydrodynamic fluctuations in heterogeneous adjacent multidomains in shear flow. Physical Review E, 93(3):033312, 2016. |
Blood Cell Modeling
The human red blood cell are vital for oxygen transportation in our circulatory system. My work on modeling red blood cells spans both the microscopic and mesoscopic scale. At the micro scale, I have worked on modeling the entire membrane and cytoskeleton of an RBC from protein resolution using a two-component coarse-grained molecular dynamics model. This type of simulation enables an array of in silico experiments such as measuring RBC shear and bending modulus in the presence of structural defects, studying uncoupling between the lipid bilayer and cytoskeleton, and modeling cellular uptake of elastic nanoparticles. At the mesoscale, I have modeled cell sorting microfluidic devices that can exploit the deterministic lateral displacement effect to separate individual circulating tumor cells from millions of surrounding red blood cells.
| [1] | Patient-specific blood rheology in sickle-cell anaemia. Interface focus, 6(1):20150065, 2016. |
| [2] | The in-silico lab-on-a-chip: petascale and high-throughput simulations of microfluidics at cell resolution. In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, 2015. |